Data Submission Walkthrough
This guide details step-by-step procedures for different aspects of the GDC Data Submission process and how they relate to the GDC Data Model and structure. The first sections of this guide break down the submission process and associate each step with the Data Model. Additional sections are detailed below for strategies on expediting data submission, using features of the GDC Data Submission Portal, and best practices used by the GDC.
GDC Data Model Basics
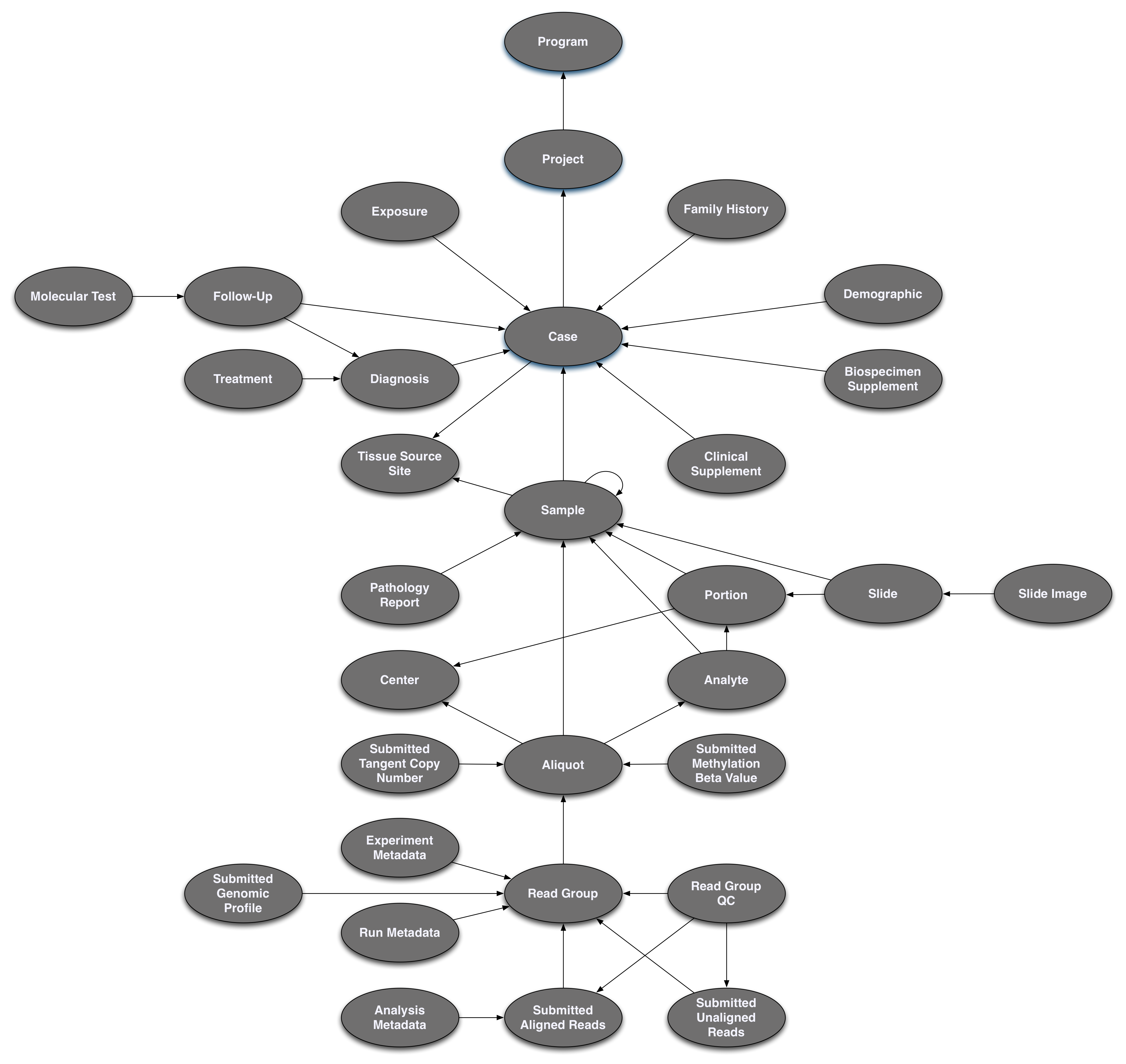
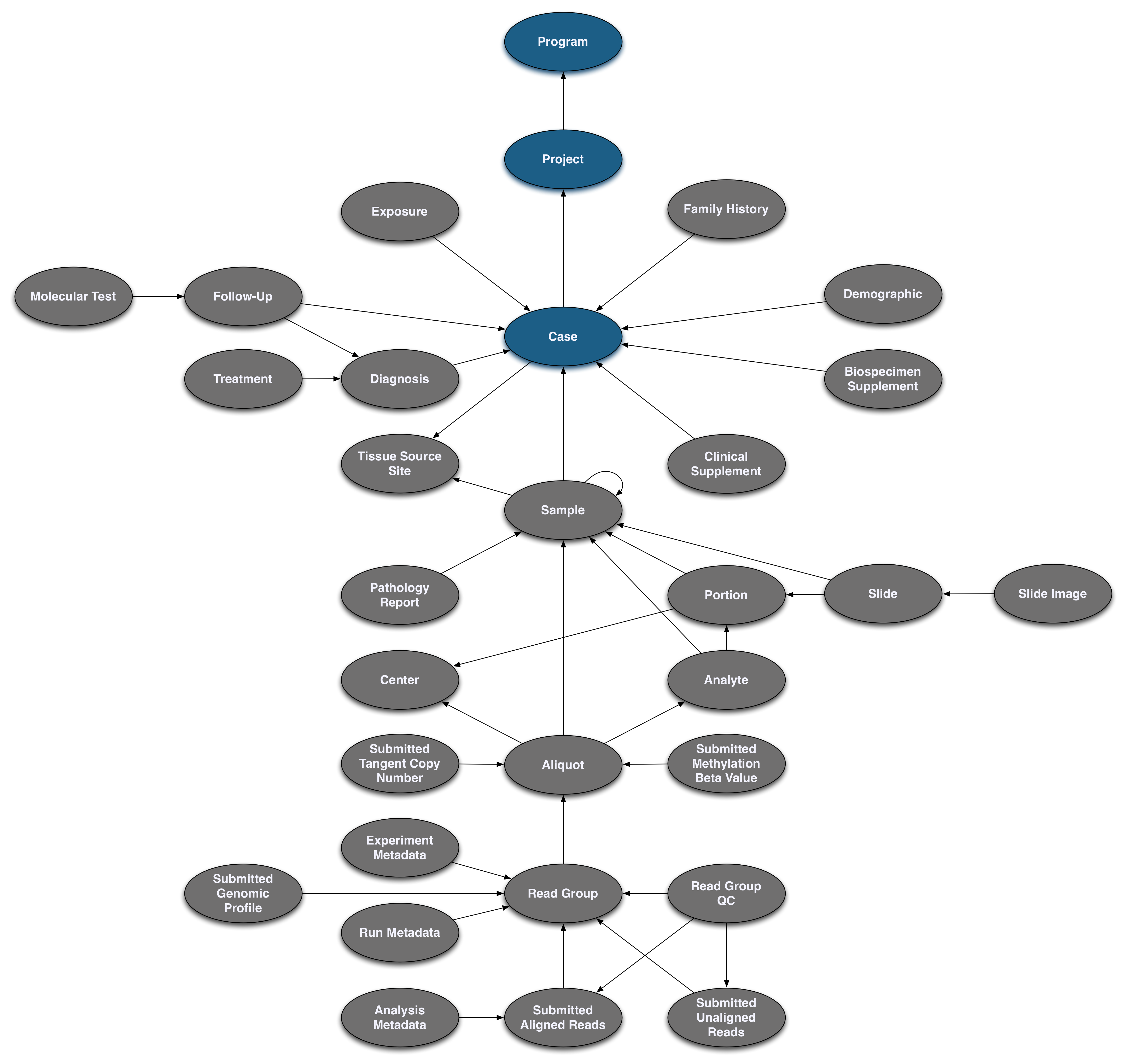
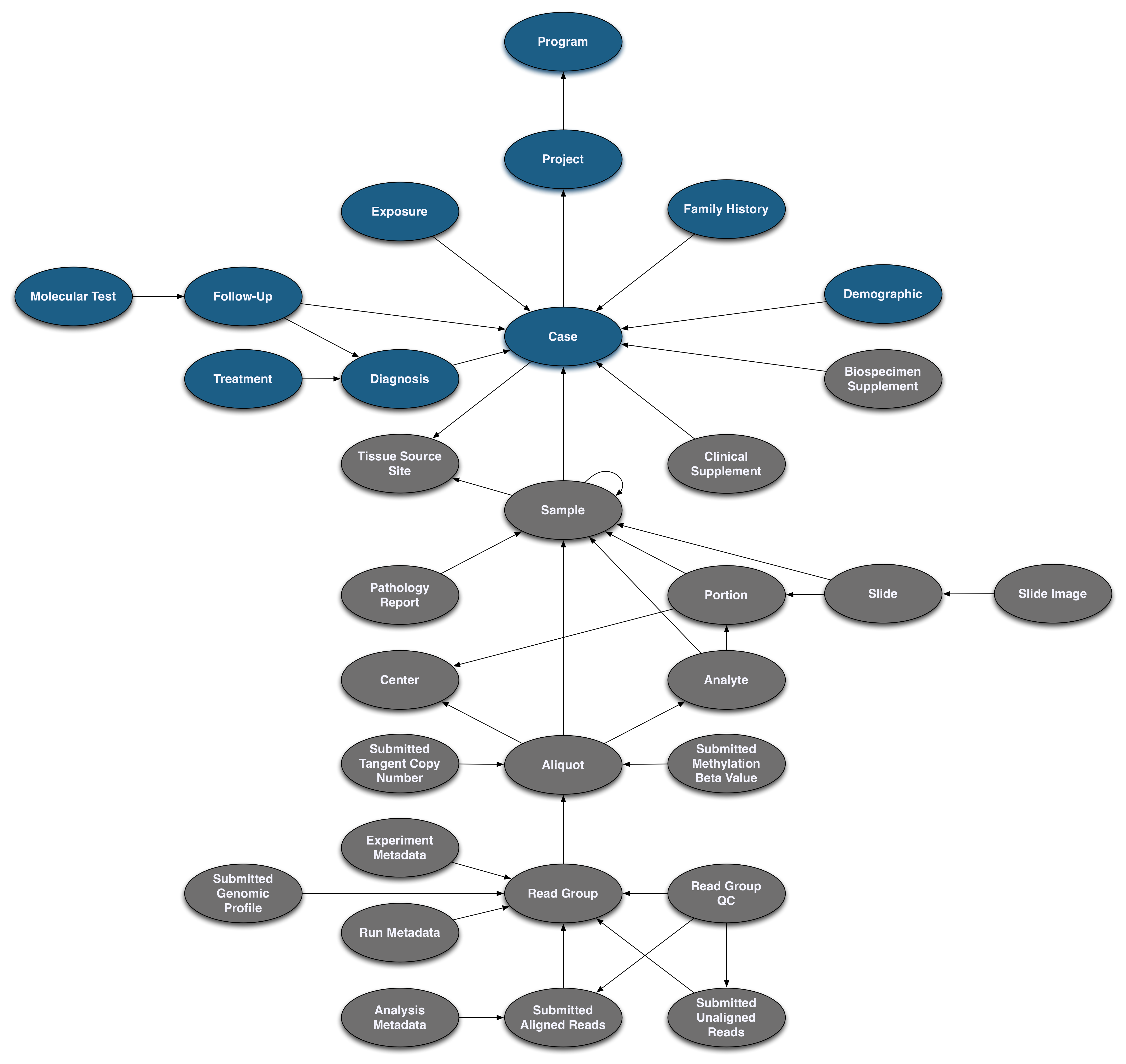
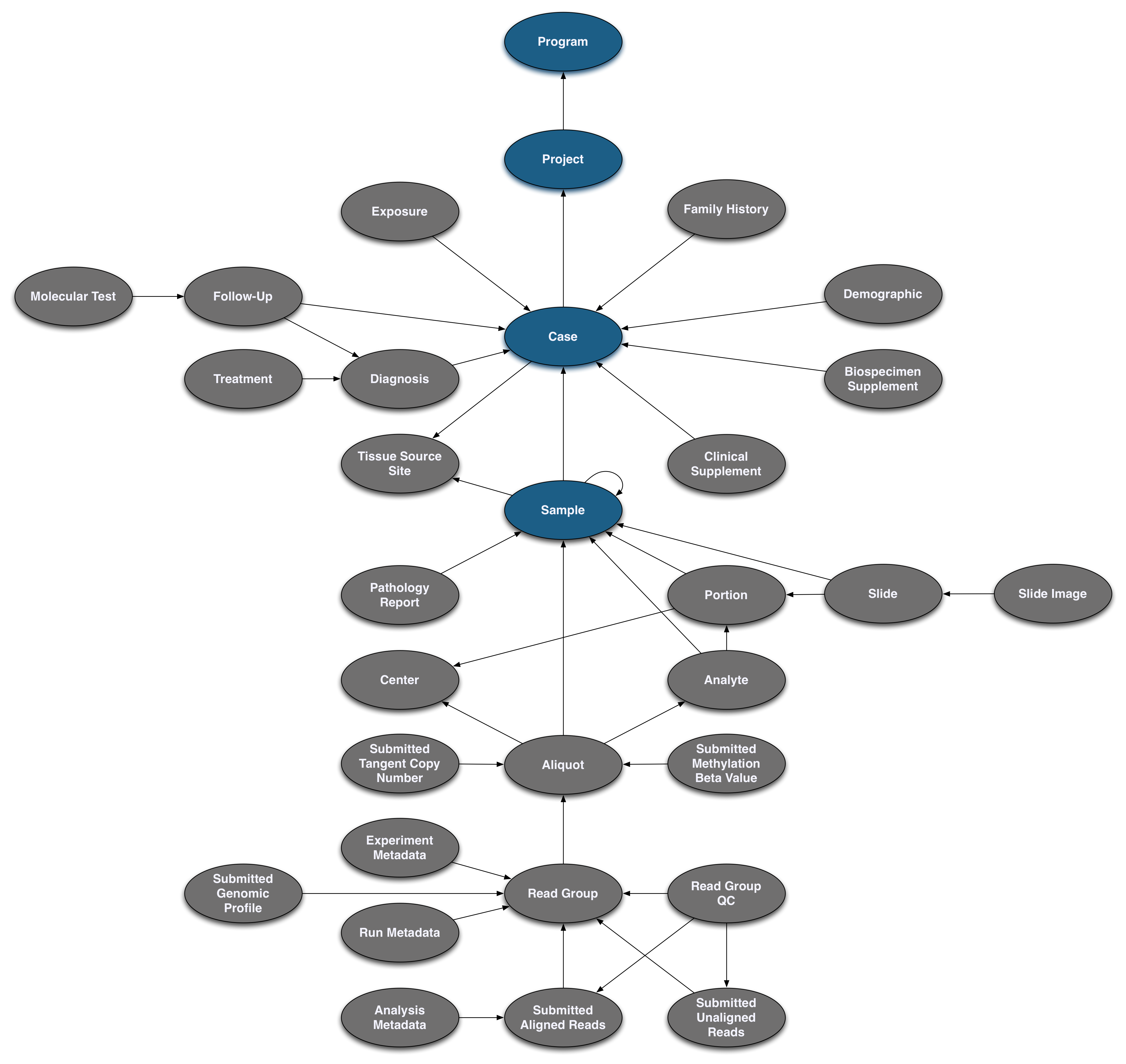
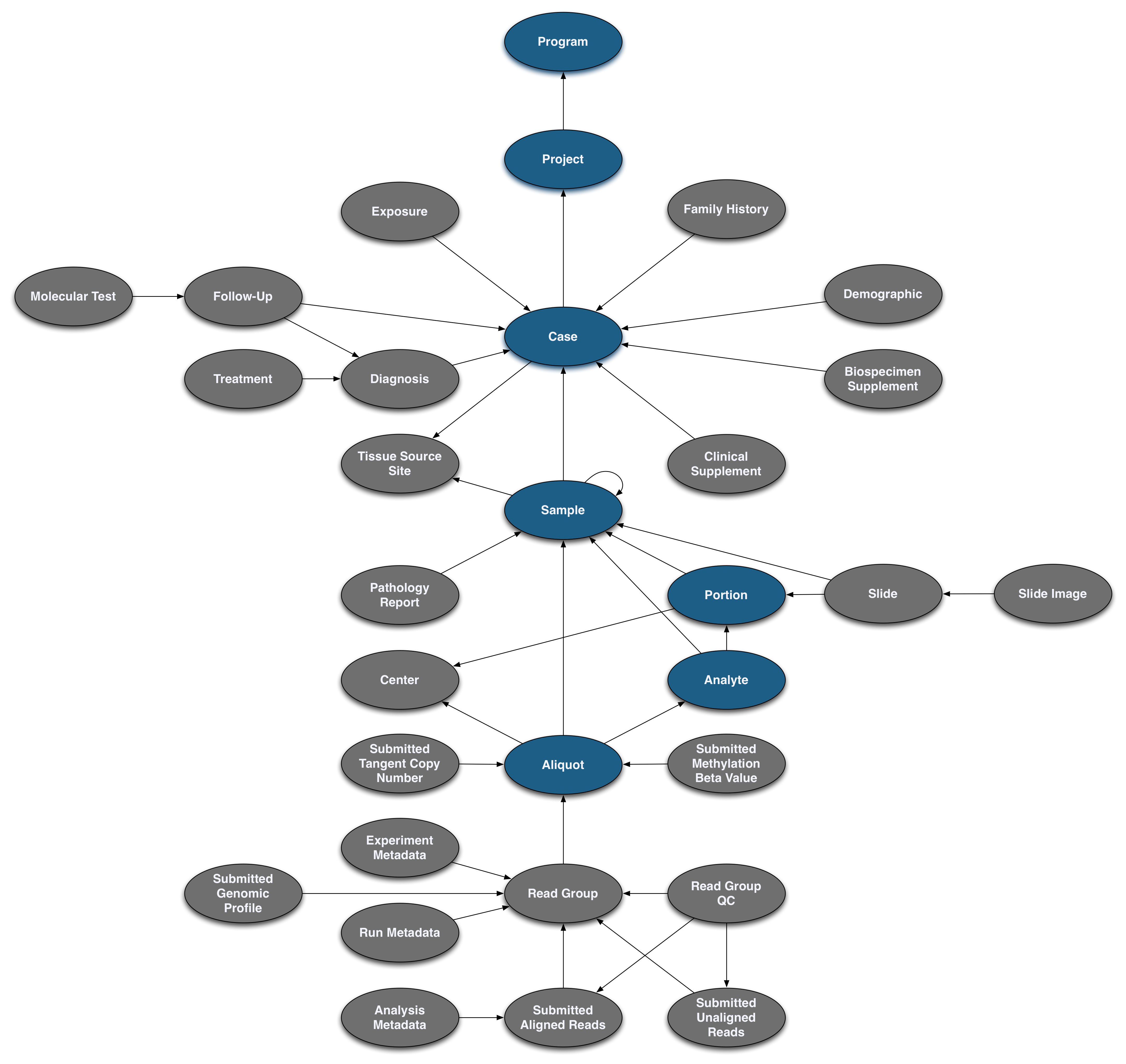
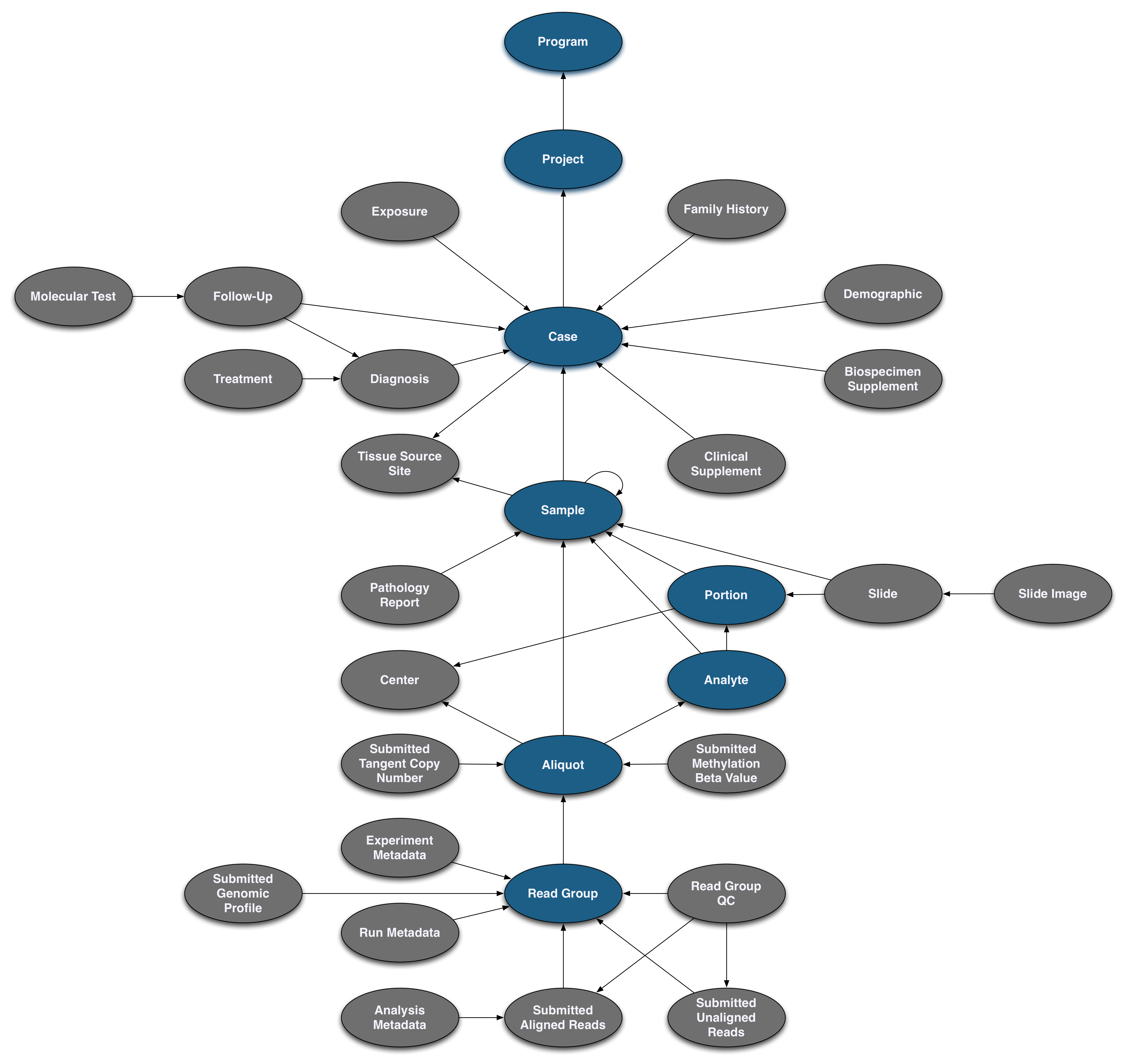
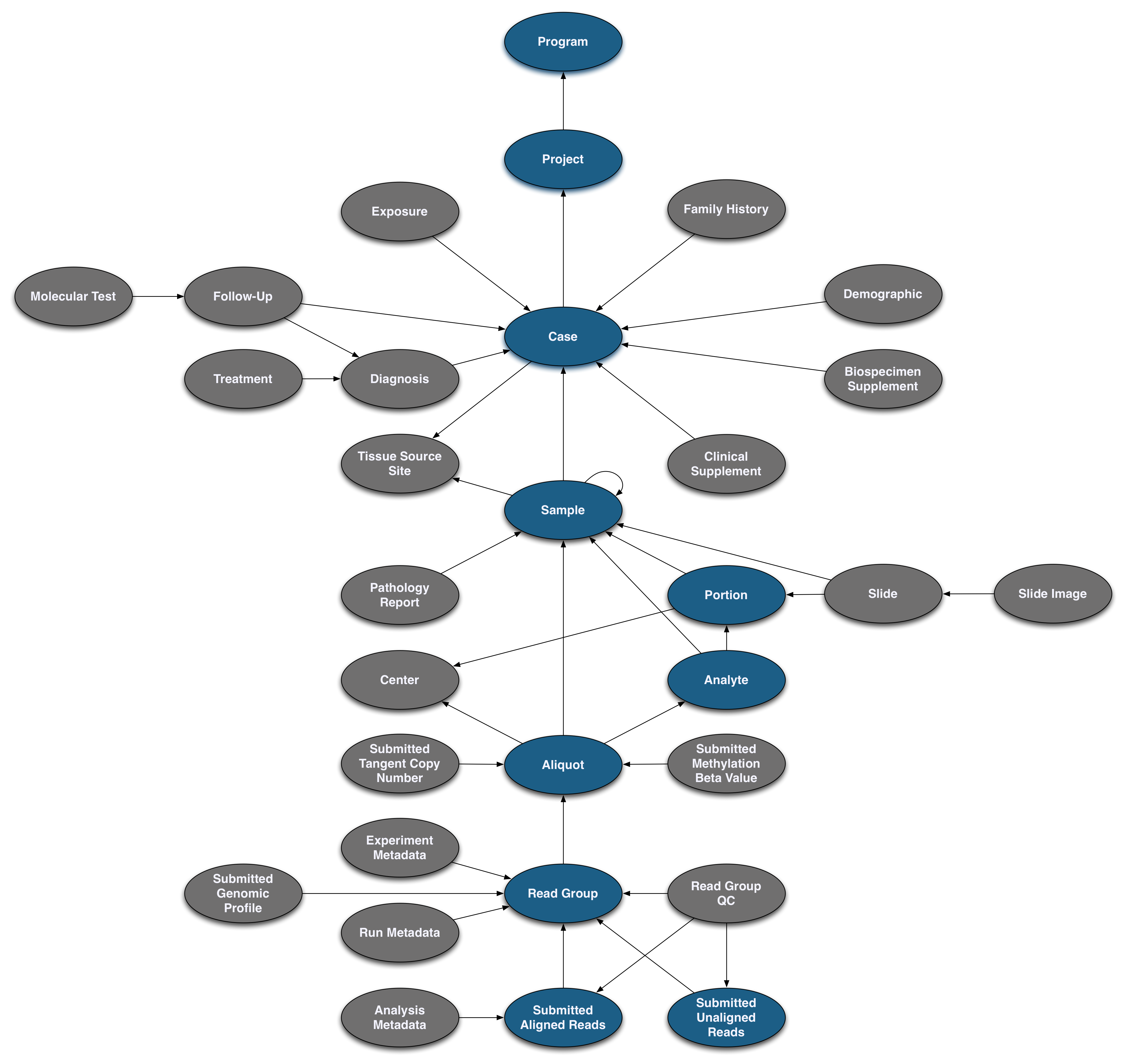
Pictured below is the submittable subset of the GDC Data Model: a roadmap for GDC data submission. Each oval node in the graphic represents an entity: a logical unit of data related to a specific clinical, biospecimen, or file facet in the GDC. An entity includes a set of fields, the associated values, and information about its related node associations. All submitted entities require a connection to another entity type, based on the GDC Data Model, and a submitter_id as an identifier. This walkthrough will go through the submission of different entities. The completed (submitted) portion of the entity process will be highlighted in blue.
Case Submission
The case is the center of the GDC Data Model and usually describes a specific patient. Each case is connected to a project. Different types of clinical data, such as diagnoses and exposures, are connected to the case to describe the case's attributes and medical information.
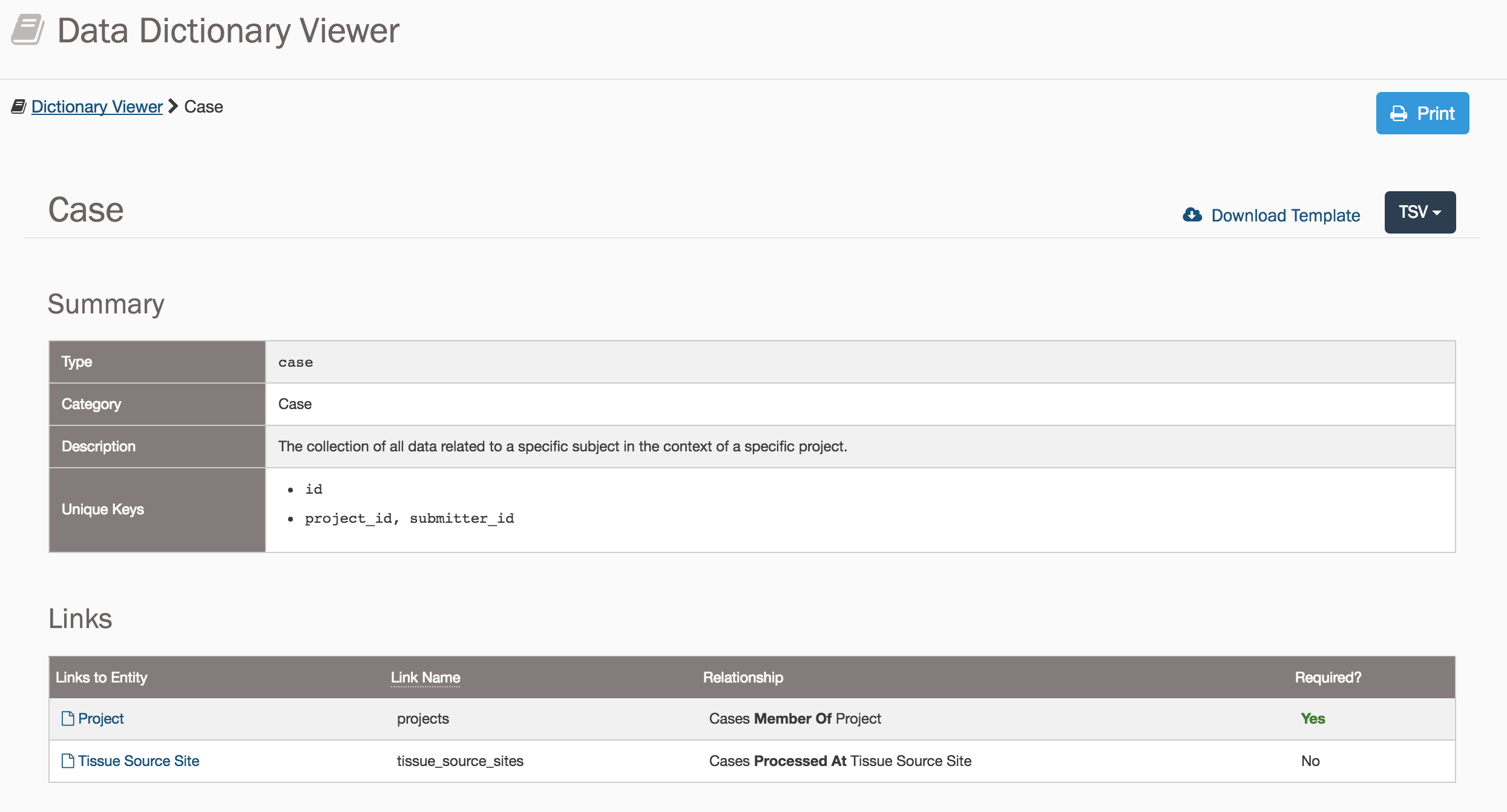
The main entity of the GDC Data Model is the case, each of which must be registered beforehand with dbGaP under a unique submitter_id. The first step to submitting a case is to consult the Data Dictionary, which details the fields that are associated with a case, the fields that are required to submit a case, and the values that can populate each field. Dictionary entries are available for all entities in the GDC Data Model.
Submitting a Case entity requires:
submitter_id: A unique key to identify thecaseprojects.code: A link to theproject
The submitter ID is different from the universally unique identifier (UUID), which is based on the UUID Version 4 Naming Convention. The UUID can be accessed under the <entity_type>_id field for each entity. For example, the case UUID can be accessed under the case_id field. The UUID is either assigned to each entity automatically or can be submitted by the user. Submitter-generated UUIDs cannot be uploaded in submittable_data_file entity types. See the Data Model Users Guide for more details about GDC identifiers.
The projects.code field connects the case entity to the project entity. The rest of the entity connections use the submitter_id field instead.
The case entity can be added in JSON or TSV format. A template for any entity in either of these formats can be found in the Data Dictionary at the top of each page. Templates populated with case metadata in both formats are displayed below.
{
"type": "case",
"submitter_id": "PROJECT-INTERNAL-000055",
"projects": {
"code": "INTERNAL"
}
}
type submitter_id projects.code
case PROJECT-INTERNAL-000055 INTERNAL
Note: JSON and TSV formats handle links between entities (
caseandproject) differently. JSON includes thecodefield nested withinprojectswhile TSV appendscodetoprojectswith a period.
Uploading the Case Submission File
The file detailed above can be uploaded using the GDC Data Submission Portal and the GDC API as described below:
Upload Using the GDC Data Submission Portal
An example of a case upload is detailed below. The GDC Data Submission Portal is equipped with a wizard window to facilitate the upload and validation of entities.
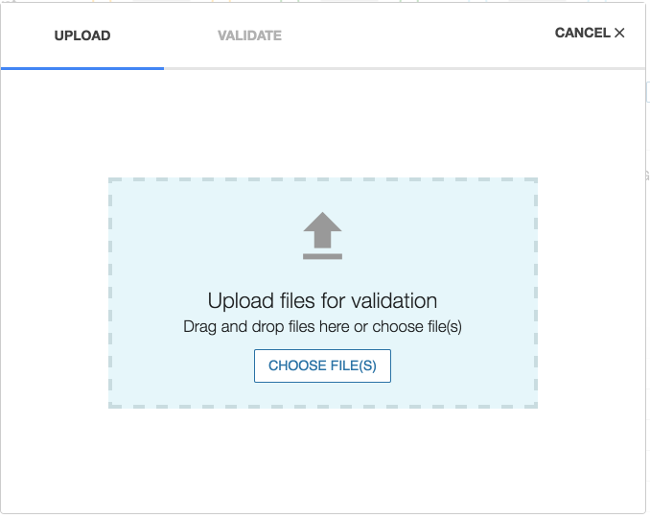
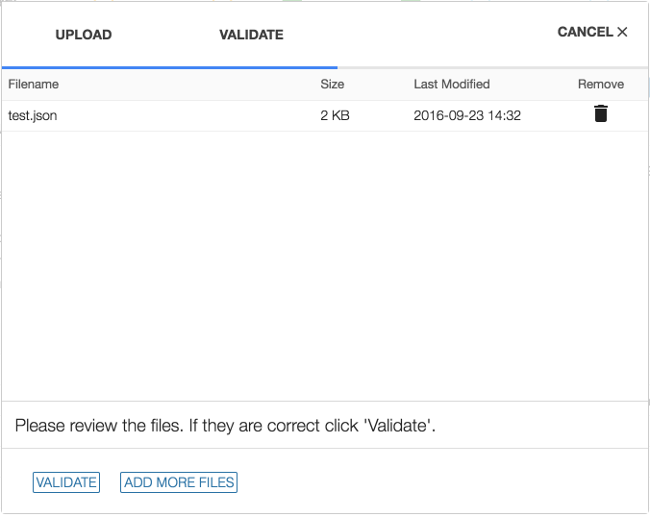
1. Upload Files
Choosing 'UPLOAD' from the project dashboard will open the Upload Data Wizard.
Files containing one or more entities can be added either by clicking on CHOOSE FILE(S) or using drag and drop. Files can be removed from the Upload Data Wizard by clicking on the garbage can icon that is displayed next to the file after the file is selected for upload.
2. Validate Entities
The Validate Entities stage acts as a safeguard against submitting incorrectly formatted data to the GDC Data Submission Portal. During the validation stage, the GDC API will validate the content of uploaded entities against the Data Dictionary to detect potential errors. Invalid entities will not be processed and must be corrected by the user and re-uploaded before being accepted. A validation error report provided by the system can be used to isolate and correct errors.
When the first file is added, the wizard will move to the Validate section and the user can continue to add files. When all files have been added, choosing VALIDATE will run a test to check if the entities are valid for submission.
3. Commit or Discard Files
If the upload contains valid entities, a new transaction will appear in the latest transactions panel with the option to COMMIT or DISCARD the data. Entities contained in these files can be committed (applied) to the project or discarded using these two buttons.
If the upload contains invalid files, a transaction will appear with a FAILED status. Invalid files will need to be either corrected and re-uploaded or removed from the submission. If more than one file is uploaded and at least one is not valid, the validation step will fail for all files.
Upload Using the GDC API
The API has a much broader range of functionality than the Data Wizard. Entities can be created, updated, and deleted through the API. See the API Submission User Guide for a more detailed explanation and for the rest of the functionalities of the API. Generally, uploading an entity through the API can be performed using a command similar to the following:
curl --header "X-Auth-Token: $token" --request POST --data @CASE.json https://api.gdc.cancer.gov/v0/submission/GDC/INTERNAL/_dry_run?async=true
CASE.json is detailed below.
{
"type": "case",
"submitter_id": "PROJECT-INTERNAL-000055",
"projects": {
"code": "INTERNAL"
}
}
In this example, the _dry_run marker is used to determine if the entities can be validated, but without committing any information. If a command passed through the _dry_run works, the command will work when it is changed to commit. For more information please go to Dry Run Transactions.
Note: Submission of TSV files is also supported by the GDC API.
Next, the file can either be committed (applied to the project) through the Data Submission Portal as before, or another API query can be performed that will commit the file to the project. The transaction number in the URL (467) is printed to the console during the first step of API submission and can also be retrieved from the Transactions tab in the Data Submission Portal.
curl --header "X-Auth-Token: $token" --request POST https://api.gdc.cancer.gov/v0/submission/GDC/INTERNAL/transactions/467/commit?async=true
Clinical Data Submission
Typically, a submission project will include additional information about a case such as demographic, diagnosis, or exposure data.
Clinical Data Requirements
For the GDC to release a project there is a minimum number of clinical properties that are required. Minimal GDC requirements for each project includes age, gender, and diagnosis information. Other requirements may be added when the submitter is approved for submission to the GDC.
Submitting a Demographic Entity to a Case
The demographic entity contains information that characterizes the case entity.
Submitting a Demographic entity requires:
submitter_id: A unique key to identify thedemographicentity.cases.submitter_id: The unique key that was used for thecasethat links thedemographicentity to thecase.ethnicity: An individual's self-described social and cultural grouping, specifically whether an individual describes themselves as Hispanic or Latino. The provided values are based on the categories defined by the U.S. Office of Management and Business and used by the U.S. Census Bureau.gender: Text designations that identify gender. Gender is described as the assemblage of properties that distinguish people on the basis of their societal roles.race: An arbitrary classification of a taxonomic group that is a division of a species. It usually arises as a consequence of geographical isolation within a species and is characterized by shared heredity, physical attributes and behavior, and in the case of humans, by common history, nationality, or geographic distribution. The provided values are based on the categories defined by the U.S. Office of Management and Business and used by the U.S. Census Bureau.
{
"type": "demographic",
"submitter_id": "PROJECT-INTERNAL-000055-DEMOGRAPHIC-1",
"cases": {
"submitter_id": "PROJECT-INTERNAL-000055"
},
"ethnicity": "not hispanic or latino",
"gender": "male",
"race": "asian",
}
type cases.submitter_id ethnicity gender race
demographic PROJECT-INTERNAL-000055 not hispanic or latino male asian
Submitting a Diagnosis Entity to a Case
Submitting a Diagnosis entity requires:
submitter_id: A unique key to identify thediagnosisentity.cases.submitter_id: The unique key that was used for thecasethat links thediagnosisentity to thecase.age_at_diagnosis: Age at the time of diagnosis expressed in number of days since birth.days_to_last_follow_up: Time interval from the date of last follow up to the date of initial pathologic diagnosis, represented as a calculated number of days.days_to_last_known_disease_status: Time interval from the date of last follow up to the date of initial pathologic diagnosis, represented as a calculated number of days.days_to_recurrence: Time interval from the date of new tumor event including progression, recurrence and new primary malignancies to the date of initial pathologic diagnosis, represented as a calculated number of days.last_known_disease_status: The state or condition of an individual's neoplasm at a particular point in time.morphology: The third edition of the International Classification of Diseases for Oncology, published in 2000 used principally in tumor and cancer registries for coding the site (topography) and the histology (morphology) of neoplasms. The study of the structure of the cells and their arrangement to constitute tissues and, finally, the association among these to form organs. In pathology, the microscopic process of identifying normal and abnormal morphologic characteristics in tissues, by employing various cytochemical and immunocytochemical stains. A system of numbered categories for representation of data.primary_diagnosis: Text term for the structural pattern of cancer cells used to define a microscopic diagnosis.progression_or_recurrence: Yes/No/Unknown indicator to identify whether a patient has had a new tumor event after initial treatment.site_of_resection_or_biopsy: The third edition of the International Classification of Diseases for Oncology, published in 2000, used principally in tumor and cancer registries for coding the site (topography) and the histology (morphology) of neoplasms. The description of an anatomical region or of a body part. Named locations of, or within, the body. A system of numbered categories for representation of data.tissue_or_organ_of_origin: Text term that describes the anatomic site of the tumor or disease.tumor_grade: Numeric value to express the degree of abnormality of cancer cells, a measure of differentiation and aggressiveness.tumor_stage: The extent of a cancer in the body. Staging is usually based on the size of the tumor, whether lymph nodes contain cancer, and whether the cancer has spread from the original site to other parts of the body. The accepted values for tumor_stage depend on the tumor site, type, and accepted staging system. These items should accompany the tumor_stage value as associated metadata.vital_status: The survival state of the person registered on the protocol.
{
"type": "diagnosis",
"submitter_id": "PROJECT-INTERNAL-000055-DIAGNOSIS-1",
"cases": {
"submitter_id": "GDC-INTERNAL-000099"
},
"age_at_diagnosis": 10256,
"days_to_last_follow_up": 34,
"days_to_last_known_disease_status": 34,
"days_to_recurrence": 45,
"last_known_disease_status": "Tumor free",
"morphology": "8260/3",
"primary_diagnosis": "ACTH-producing tumor",
"progression_or_recurrence": "no",
"site_of_resection_or_biopsy": "Lung, NOS",
"tissue_or_organ_of_origin": "Lung, NOS",
"tumor_grade": "Not Reported",
"tumor_stage": "stage i",
"vital_status": "alive"
}
type submitter_id cases.submitter_id age_at_diagnosis days_to_last_follow_up days_to_last_known_disease_status days_to_recurrence last_known_disease_status morphology primary_diagnosis progression_or_recurrence site_of_resection_or_biopsy tissue_or_organ_of_origin tumor_grade tumor_stage vital_status
diagnosis PROJECT-INTERNAL-000055-DIAGNOSIS-1 GDC-INTERNAL-000099 10256 34 34 45 Tumor free 8260/3 ACTH-producing tumor no Lung, NOS Lung, NOS not reported stage i alive
Submitting an Exposure Entity to a Case
Submitting an Exposure entity does not require any information besides a link to the case and a submitter_id. The following fields are optionally included:
alcohol_history: A response to a question that asks whether the participant has consumed at least 12 drinks of any kind of alcoholic beverage in their lifetime.alcohol_intensity: Category to describe the patient's current level of alcohol use as self-reported by the patient.alcohol_days_per_week: Numeric value used to describe the average number of days each week that a person consumes an alchoolic beverage.years_smoked: Numeric value (or unknown) to represent the number of years a person has been smoking.tobacco_smoking_onset_year: The year in which the participant began smoking.tobacco_smoking_quit_year: The year in which the participant quit smoking.
{
"type": "exposure",
"submitter_id": "PROJECT-INTERNAL-000055-EXPOSURE-1",
"cases": {
"submitter_id": "PROJECT-INTERNAL-000055"
},
"alcohol_history": "yes",
"alcohol_intensity": "Drinker",
"alcohol_days_per_week": 2,
"years_smoked": 5,
"tobacco_smoking_onset_year": 2007,
"tobacco_smoking_quit_year": 2012
}
type submitter_id cases.submitter_id alcohol_history alcohol_intensity alcohol_days_per_week years_smoked tobacco_smoking_onset_year tobacco_smoking_quit_year
exposure PROJECT-INTERNAL-000055-EXPOSURE-1 PROJECT-INTERNAL-000055 yes Drinker 2 5 2007 2012
Note: Submitting a clinical entity uses the same conventions as submitting a
caseentity (detailed above).
Biospecimen Submission
One of the main features of the GDC is the genomic data harmonization workflow. Genomic data is connected the case through biospecimen entities. The sample entity describes a biological piece of matter that originated from a case. Subsets of the sample such as portions and analytes can optionally be described. The aliquot originates from a sample or analyte and describes the nucleic acid extract that was sequenced. The read_group entity describes the resulting set of reads from one sequencing lane.
Sample Submission
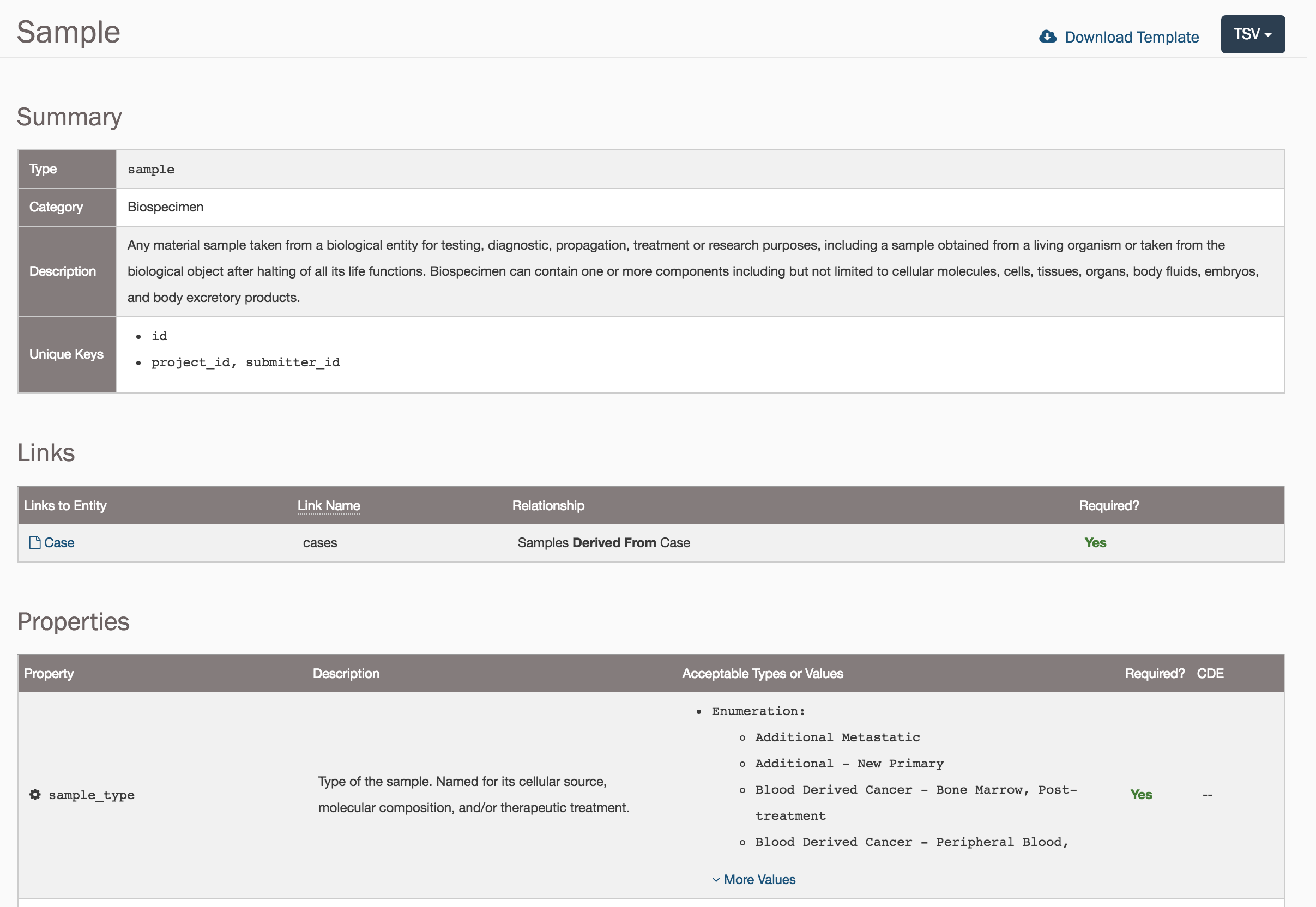
A sample submission has the same general structure as a case submission as it will require a unique key and a link to the case. However, sample entities require four additional values:
tissue_typetumor_descriptorspecimen_typepreservation_method
This peripheral data is required because it is necessary for the data to be interpreted. For example, an investigator using this data would need to know whether the sample came from tumor or normal tissue.
Submitting a Sample entity requires:
submitter_id: A unique key to identify thesample.cases.submitter_id: The unique key that was used for thecasethat links thesampleto thecase.tissue_type: Text term that represents a description of the kind of tissue collected with respect to disease status or proximity to tumor tissue.tumor_descriptor: Text that describes the kind of disease present in the tumor specimen as related to a specific timepoint.specimen_type: The type of a material sample taken from a biological entity for testing, diagnostic, propagation, treatment or research purposes. This includes particular types of cellular molecules, cells, tissues, organs, body fluids, embryos, and body excretory substances.preservation_method: Text term that represents the method used to preserve the sample.
Note: The
casemust be "committed" to the project before asamplecan be linked to it. This also applies to all other links between entities.
{
"type": "sample",
"cases": {
"submitter_id": "PROJECT-INTERNAL-000055"
},
"submitter_id": "Blood-00001SAMPLE_55",
"tissue_type": "Normal",
"tumor_descriptor": "Not Applicable",
"specimen_type": "Peripheral Blood NOS",
"preservation_method": "Frozen"
}
type cases.submitter_id submitter_id tissue_type specimen_type tumor_descriptor preservation_method
sample PROJECT-INTERNAL-000055 Blood-00001SAMPLE_55 Normal Peripheral Blood NOS Not Applicable Frozen
Portion, Analyte and Aliquot Submission
Submitting a Portion entity requires:
submitter_id: A unique key to identify theportion.samples.submitter_id: The unique key that was used for thesamplethat links theportionto thesample.
{
"type": "portion",
"submitter_id": "Blood-portion-000055",
"samples": {
"submitter_id": "Blood-00001SAMPLE_55"
}
}
type submitter_id samples.submitter_id
portion Blood-portion-000055 Blood-00001SAMPLE_55
Submitting an Analyte entity requires:
submitter_id: A unique key to identify theanalyte.portions.submitter_id: The unique key that was used for theportionthat links theanalyteto theportion.analyte_type: Text term that represents the kind of molecular specimen analyte.
{
"type": "analyte",
"portions": {
"submitter_id": "Blood-portion-000055"
},
"analyte_type": "DNA",
"submitter_id": "Blood-analyte-000055"
}
type portions.submitter_id analyte_type submitter_id
analyte Blood-portion-000055 DNA Blood-analyte-000055
Submitting an Aliquot entity requires:
submitter_id: A unique key to identify thealiquot.analytes.submitter_id: The unique key that was used for theanalytethat links thealiquotto theanalyte.
{
"type": "aliquot",
"submitter_id": "Blood-00021-aliquot55",
"analytes": {
"submitter_id": "Blood-analyte-000055"
}
}
type submitter_id analytes.submitter_id
aliquot Blood-00021-aliquot55 Blood-analyte-000055
Note:
aliquotentities can be directly linked tosampleentities via thesamples.submitter_id. Theportionandanalyteentities are not required for submission.
Read Group Submission
Information about sequencing reads is necessary for downstream analysis, thus the read_group entity requires more fields than the other Biospecimen entities (sample, portion, analyte, aliquot).
Submitting a Read Group entity requires:
submitter_id: A unique key to identify theread_group.aliquots.submitter_id: The unique key that was used for thealiquotthat links theread_groupto thealiquot.experiment_name: Submitter-defined name for the experiment.is_paired_end: Are the reads paired end? (Boolean value:trueorfalse).library_name: Name of the library.library_strategy: Library strategy.platform: Name of the platform used to obtain data.read_group_name: The name of theread_group.read_length: The length of the reads (integer).sequencing_center: Name of the center that provided the sequence files.library_selection: Library Selection Method.target_capture_kit: Description that can uniquely identify a target capture kit. Suggested value is a combination of vendor, kit name, and kit version.
{
"type": "read_group",
"submitter_id": "Blood-00001-aliquot_lane1_barcodeACGTAC_55",
"experiment_name": "Resequencing",
"is_paired_end": true,
"library_name": "Solexa-34688",
"library_strategy": "WXS",
"platform": "Illumina",
"read_group_name": "205DD.3-2",
"read_length": 75,
"sequencing_center": "BI",
"library_selection": "Hybrid Selection",
"target_capture_kit": "Custom MSK IMPACT Panel - 468 Genes",
"aliquots":
{
"submitter_id": "Blood-00021-aliquot55"
}
}
type submitter_id experiment_name is_paired_end library_name library_selection library_strategy platform read_group_name read_length sequencing_center target_capture_kit aliquots.submitter_id
read_group Blood-00001-aliquot_lane1_barcodeACGTAC_55 Resequencing true Solexa-34688 Hybrid Selection WXS Illumina 205DD.3-2 75 BI Custom MSK IMPACT Panel - 468 Genes Blood-00021-aliquot55
Note: Submitting a biospecimen entity uses the same conventions as submitting a
caseentity (detailed above).
Experiment Data Submission
Several types of experiment data can be uploaded to the GDC. The submitted_aligned_reads and submitted_unaligned_reads files are associated with the read_group entity, while the array-based files such as the submitted_tangent_copy_number are associated with the aliquot entity. Each of these file types are described in their respective entity submission and are uploaded separately using the GDC API or the GDC Data Transfer Tool.
Before the experiment data file can be submitted, the GDC requires that the user provides information about the file as a submittable_data_file entity. This includes file-specific data needed to validate the file and assess which analyses should be performed. Sequencing data files can be submitted as submitted_aligned_reads or submitted_unaligned_reads.
Submitting a Submitted Aligned-Reads (Submitted Unaligned-Reads) entity requires:
submitter_id: A unique key to identify thesubmitted_aligned_reads.read_groups.submitter_id: The unique key that was used for theread_groupthat links thesubmitted_aligned_readsto theread_group.data_category: Broad categorization of the contents of the data file.data_format: Format of the data files.data_type: Specific content type of the data file. (must be "Aligned Reads").experimental_strategy: The sequencing strategy used to generate the data file.file_name: The name (or part of a name) of a file (of any type).file_size: The size of the data file (object) in bytes.md5sum: The 128-bit hash value expressed as a 32 digit hexadecimal number used as a file's digital fingerprint.
{
"type": "submitted_aligned_reads",
"submitter_id": "Blood-00001-aliquot_lane1_barcodeACGTAC_55.bam",
"data_category": "Raw Sequencing Data",
"data_format": "BAM",
"data_type": "Aligned Reads",
"experimental_strategy": "WGS",
"file_name": "test.bam",
"file_size": 38,
"md5sum": "aa6e82d11ccd8452f813a15a6d84faf1",
"read_groups": [
{
"submitter_id": "Primary_Tumor_RG_86-1"
}
]
}
type submitter_id data_category data_format data_type experimental_strategy file_name file_size md5sum read_groups.submitter_id#1
submitted_aligned_reads Blood-00001-aliquot_lane1_barcodeACGTAC_55.bam Raw Sequencing Data BAM Aligned Reads WGS test.bam 38 aa6e82d11ccd8452f813a15a6d84faf1 Primary_Tumor_RG_86-1
Note: For details on submitting experiment data associated with more than one
read_groupentity, see the Tips for Complex Submissions section.
Uploading the Submittable Data File to the GDC
The submittable data file can be uploaded when it is registered with the GDC. A submittable data file is registered when its corresponding entity (e.g. submitted_unaligned_reads) is uploaded and committed. It is important to note that the Harmonization process does not occur on these submitted files until the user clicks the Request Submission button. Uploading the file can be performed with either the GDC Data Transfer Tool or the GDC API. Other types of data files such as clinical supplements, biospecimen supplements, and pathology reports are uploaded to the GDC in the same way. Supported data file formats are listed at the GDC Data Dictionary.
GDC Data Transfer Tool: A file can be uploaded using its UUID (which can be retrieved from the GDC Submission Portal or API) once it is registered.
The following command can be used to upload the file:
gdc-client upload --project-id PROJECT-INTERNAL --identifier a053fad1-adc9-4f2d-8632-923579128985 -t $token -f $path_to_file
Additionally a manifest can be downloaded from the Submission Portal and passed to the Data Transfer Tool. This will allow for the upload of more than one submittable_data_file:
gdc-client upload -m manifest.yml -t $token
API Upload: A submittable_data_file can be uploaded through the API by using the /submission/$PROGRAM/$PROJECT/files endpoint. The following command would be typically used to upload a file:
curl --request PUT --header "X-Auth-Token: $token" https://api.gdc.cancer.gov/v0/submission/PROJECT/INTERNAL/files/6d45f2a0-8161-42e3-97e6-e058ac18f3f3 -d $path_to_file
For more details on how to upload a submittable_data_file to a project see the API Users Guide and the Data Transfer Tool Users Guide.
Annotation Submission
The GDC Data Portal supports the use of annotations for any submitted entity or file. An annotation entity may include comments about why particular patients or samples are not present or why they may exhibit critical differences from others. Annotations include information that cannot be submitted to the GDC through other existing nodes or properties.
If a submitter would like to create an annotation, please contact the GDC Support Team (support@nci-gdc.datacommons.io).
Deleting Submitted Entities
The GDC Data Submission Portal allows users to delete submitted entities from the project when the project is in an "OPEN" state. Files cannot be deleted while in the "SUBMITTED" state. This section applies to entities that have been committed to the project. Entities that have not been committed can be removed from the project by choosing the DISCARD button. Entities can also be deleted using the API. See the API Submission Documentation for specific instructions.
NOTE: Entities associated with files uploaded to the GDC object store cannot be deleted until the associated file has been deleted. Users must utilize the GDC Data Transfer Tool to delete these files first.
Simple Deletion
If an entity was uploaded and has no related entities, it can be deleted from the Browse tab. Once the entity to be deleted is selected, choose the DELETE button in the right panel under "ACTIONS".
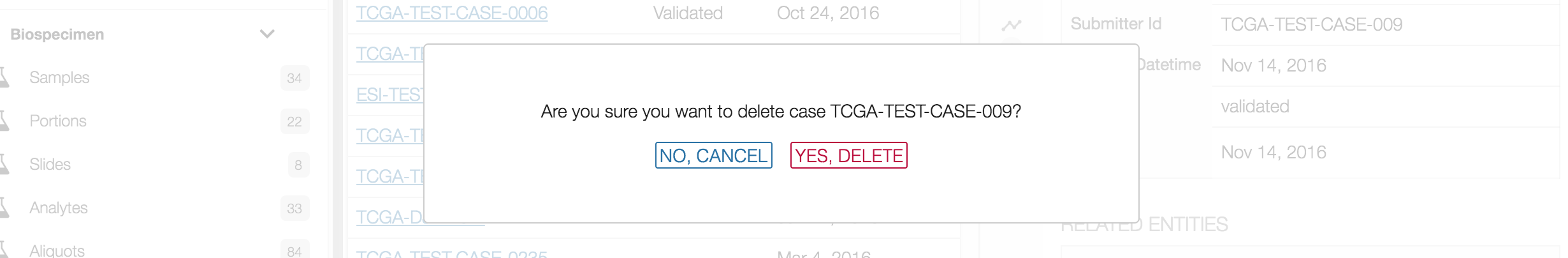
A message will then appear asking if you are sure about deleting the entity. Choosing the YES, DELETE button will remove the entity from the project, whereas choosing the NO, CANCEL button will return the user to the previous screen.
Deletion with Dependents
If an entity has related entities, such as a case with multiple samples and aliquots, deletion takes one extra step.
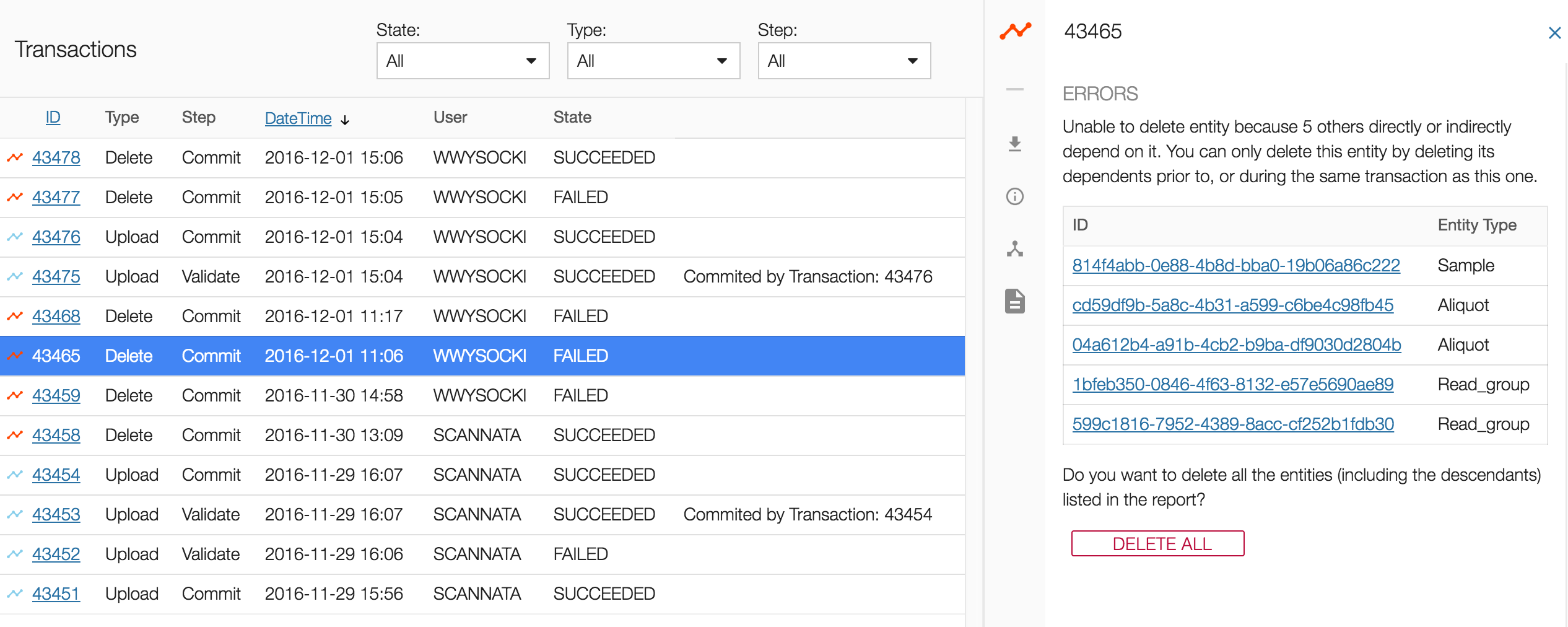
Follow the Simple Deletion method until the end. This action will appear in the Transactions tab as "Delete" with a "FAILED" state.
Choose the failed transaction and the right panel will show the list of entities related to the entity that was going to be deleted.
Selecting the DELETE ALL button at the bottom of the list will delete all of the related entities, their descendants, and the original entity.
Submitted Data File Deletion
The submittable_data_files that were uploaded erroneously are deleted separately from their associated entity using the GDC Data Transfer Tool. See the section on Deleting Data Files in the Data Transfer Tool users guide for specific instructions.
Updating Uploaded Entities
Before harmonization occurs, entities can be modified to update, add, or delete information. These methods are outlined below.
Updating or Adding Fields
Updated or additional fields can be applied to entities by re-uploading them through the GDC Data Submission portal or API. See below for an example of a case upload with a primary_site field being added and a disease_type field being updated.
{
"type":"case",
"submitter_id":"GDC-INTERNAL-000043",
"projects":{
"code":"INTERNAL"
},
"disease_type": "Myomatous Neoplasms"
}
{
"type":"case",
"submitter_id":"GDC-INTERNAL-000043",
"projects":{
"code":"INTERNAL"
},
"disease_type": "Myxomatous Neoplasms",
"primary_site": "Pancreas"
}
Guidelines:
- The newly uploaded entity must contain the
submitter_idof the existing entity so that the system updates the correct one. - All newly updated entities will be validated by the GDC Dictionary. All required fields must be present in the newly updated entity.
- Fields that are not required do not need to be re-uploaded and will remain unchanged in the entity unless they are updated.
Deleting Optional Fields
It may be necessary to delete fields from uploaded entities. This can be performed through the API and can only be applied to optional fields. It also requires the UUID of the entity, which can be retrieved from the submission portal or using a GraphQL query.
In the example below, the primary_site and disease_type fields are removed from a case entity:
curl --header "X-Auth-Token: $token_string" --request DELETE --header "Content-Type: application/json" "https://api.gdc.cancer.gov/v0/submission/EXAMPLE/PROJECT/entities/7aab7578-34ff-5651-89bb-57aefdc4c4f8?fields=primary_site,disease_type"
{
"type":"case",
"submitter_id":"GDC-INTERNAL-000043",
"projects":{
"code":"INTERNAL"
},
"disease_type": "Germ Cell Neoplasms",
"primary_site": "Pancreas"
}
{
"type":"case",
"submitter_id":"GDC-INTERNAL-000043",
"projects":{
"code":"INTERNAL"
}
}
Versioning
Changes to entities will create versions. For more information on this, please go to Uploading New Versions of Data Files.
Strategies for Submitting in Bulk
Each submission in the previous sections was broken down by component to demonstrate the GDC Data Model structure. However, the submission of multiple entities at once is supported and encouraged. Here two strategies for submitting data in an efficient manner are discussed.
Registering a BAM File: One Step
Registering a BAM file (or any other type) can be performed in one step by including all of the entities, from case to submitted_aligned_reads, in one file. See the example below:
[{
"type": "case",
"submitter_id": "PROJECT-INTERNAL-000055",
"projects": {
"code": "INTERNAL"
}
},
{
"type": "sample",
"cases": {
"submitter_id": "PROJECT-INTERNAL-000055"
},
"submitter_id": "Blood-00001SAMPLE_55",
"tissue_type": "Normal",
"tumor_descriptor": "Not Applicable",
"specimen_type": "Peripheral Blood NOS",
"preservation_method": "Frozen"
},
{
"type": "portion",
"submitter_id": "Blood-portion-000055",
"samples": {
"submitter_id": "Blood-00001SAMPLE_55"
}
},
{
"type": "analyte",
"portions": {
"submitter_id": "Blood-portion-000055"
},
"analyte_type": "DNA",
"submitter_id": "Blood-analyte-000055"
},
{
"type": "aliquot",
"submitter_id": "Blood-00021-aliquot55",
"analytes": {
"submitter_id": "Blood-analyte-000055"
}
},
{
"type": "read_group",
"submitter_id": "Blood-00001-aliquot_lane1_barcodeACGTAC_55",
"experiment_name": "Resequencing",
"is_paired_end": true,
"library_name": "Solexa-34688",
"library_selection":"Hybrid Selection",
"library_strategy": "WXS",
"platform": "Illumina",
"read_group_name": "205DD.3-2",
"read_length": 75,
"sequencing_center": "BI",
"aliquots":
{
"submitter_id": "Blood-00021-aliquot55"
}
},
{
"type": "submitted_aligned_reads",
"submitter_id": "Blood-00001-aliquot_lane1_barcodeACGTAC_55.bam",
"data_category": "Raw Sequencing Data",
"data_format": "BAM",
"data_type": "Aligned Reads",
"experimental_strategy": "WGS",
"file_name": "test.bam",
"file_size": 38,
"md5sum": "aa6e82d11ccd8452f813a15a6d84faf1",
"read_groups": [
{
"submitter_id": "Blood-00001-aliquot_lane1_barcodeACGTAC_55"
}
]
}]
All of the entities are placed into a JSON list object:
[{"type": "case","submitter_id": "PROJECT-INTERNAL-000055","projects": {"code": "INTERNAL"}}}, entity-2, entity-3]
The entities need not be in any particular order as they are validated together.
Note: Tab-delimited format is not recommended for 'one-step' submissions due to an inability of the format to accommodate multiple 'types' in one row.
Submitting Numerous Cases
The GDC understands that submitters will have projects that comprise more entities than would be reasonable to individually parse into JSON formatted files. Additionally, many investigators store large amounts of data in a tab-delimited format (TSV). For instances like this, we recommend parsing all entities of the same type into separate TSVs and submitting them on a type-basis.
For example, a user may want to submit 100 Cases associated with 100 samples, 100 portions, 100 analytes, 100 aliquots, and 100 read_groups. Constructing and submitting 100 JSON files would be tedious and difficult to organize. The solution is submitting one case TSV containing the 100 cases, one sample TSV containing the 100 samples, so on and so forth. Doing this would only require six TSVs and these files can be formatted in programs such as Microsoft Excel or Google Spreadsheets.
See the following example TSV files:
Download Previously Uploaded Metadata Files
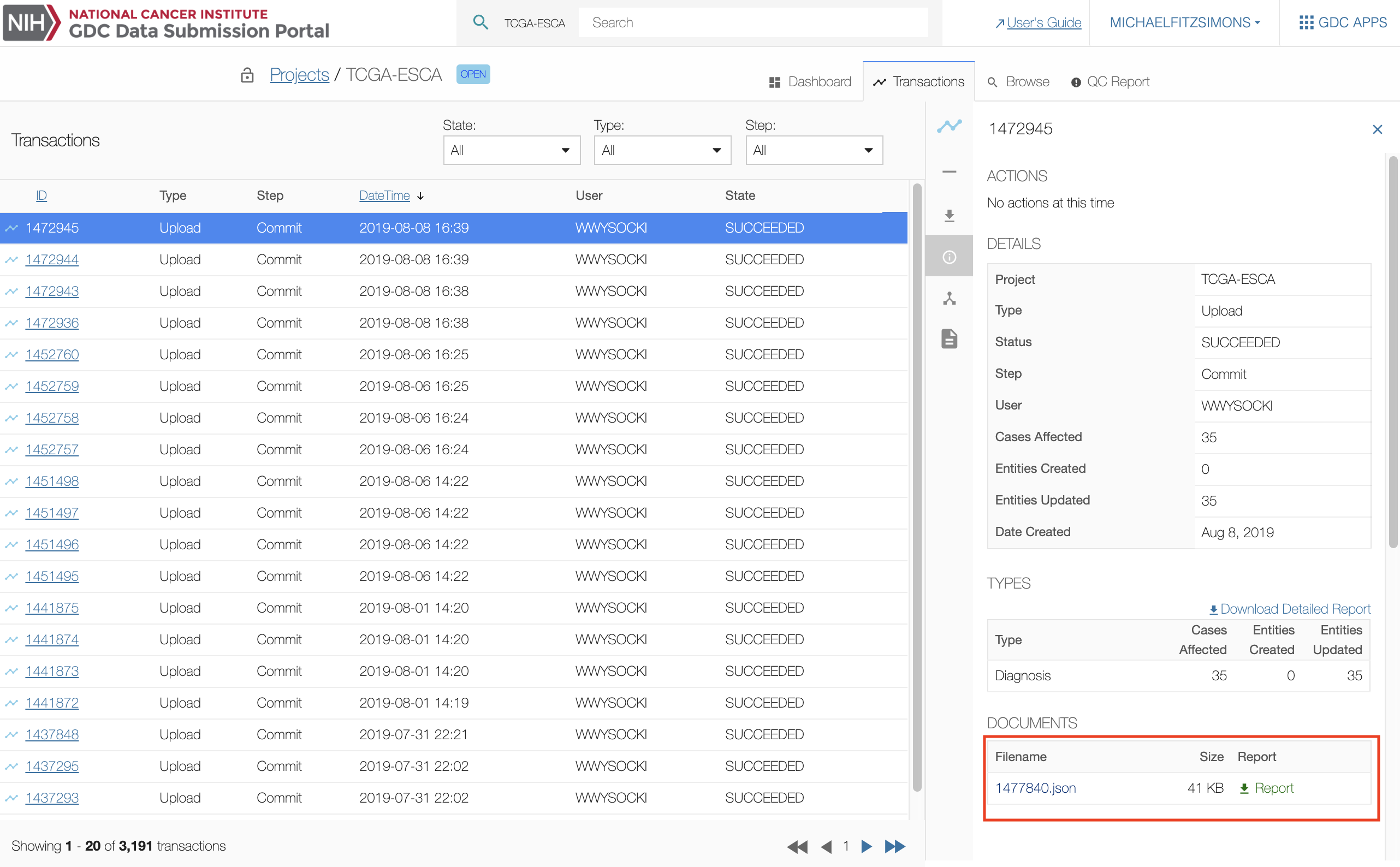
The transaction page lists all previous transactions in the project. The user can download metadata files uploaded to the GDC workspace in the details section of the screen by selecting one transaction and scrolling to the "DOCUMENTS" section.
Download Previously Uploaded Data Files
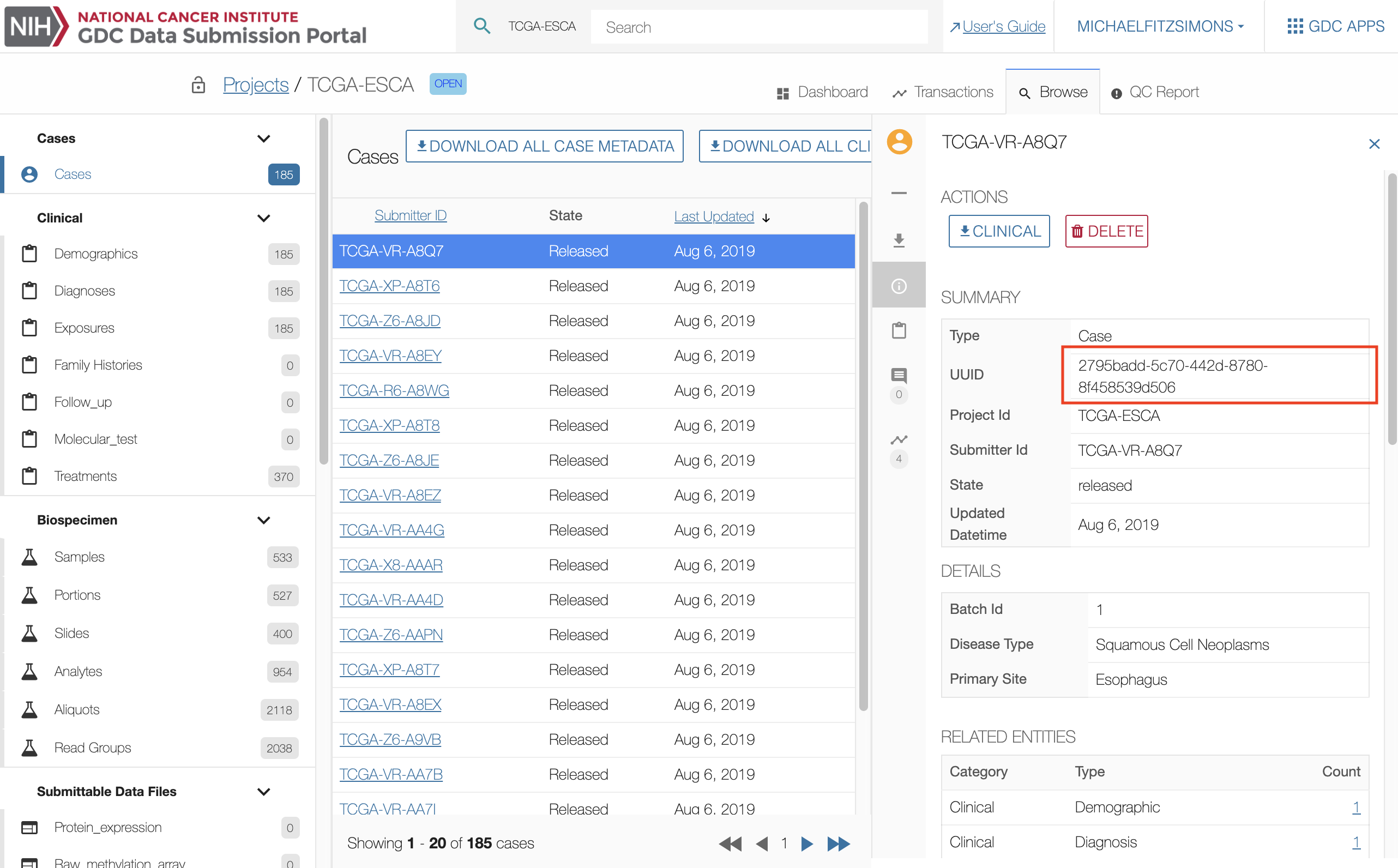
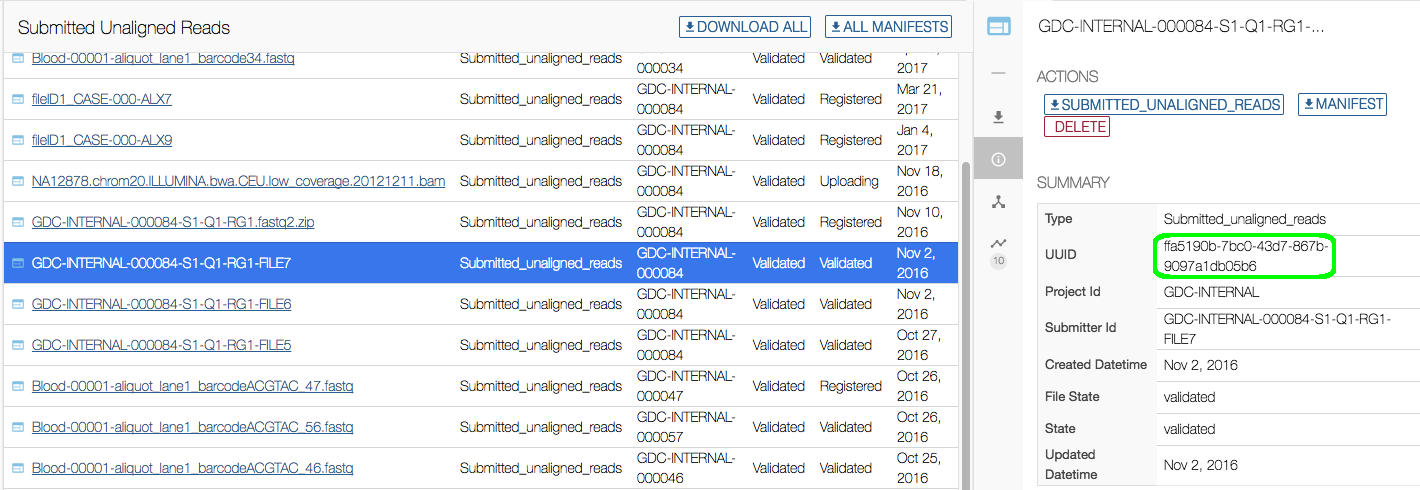
The only supported method to download data files previously uploaded to the GDC Submission Portal that have not been release yet is to use the API or the Data Transfer Tool. To retrieve data previous upload to the submission portal you will need to retrieve the data file's UUID. The UUIDs for submitted data files are located in the submission portal under the file's Summary section as well as the manifest file located on the file's Summary page.
Once the UUID(s) have been retrieved, the download process is the same as it is for downloading data files at the GDC Portal using UUIDs.
Note: When submittable data files are uploaded through the Data Transfer Tool they are not displayed as transactions.